A fresh approach to scientific engagement within the NIEHS Division of Intramural Research (DIR) began recently with the launch of the DIR Scientific Director’s Seminar Series. Inaugural lectures were filled to capacity, attended by researchers across the institute eager to expand their knowledge and collaborate.
“Discovering Gene-Environment Interactions” was presented by Alison Motsinger-Reif, Ph.D., Sept. 25, and Paul Wade, Ph.D., discussed “Epigenetics: From Mechanism to Disease Risk” Oct. 9. Motsinger-Reif is chief of the NIEHS Biostatistics and Computational Biology Branch, and Wade leads the institute’s Epigenetics and Stem Cell Biology Laboratory. Epigenetics refers to modifications to DNA or the proteins associated with DNA that can affect gene expression without altering the genetic code.

“This new lecture series will promote meaningful conversations in the Division of Intramural Research about novel scientific questions, approaches, and technologies,” said NIEHS Scientific Director Darryl Zeldin, M.D. “Seminars and Q&A sessions will be multidisciplinary, helping to spark dialogue and new collaborations that strengthen NIEHS research.”
Expanding the All of Us Program
Motsinger-Reif described her work using machine learning and other computational tools to uncover how the interplay of genes and environmental exposures can influence health. Among her other efforts, she provides leadership on the NIEHS Personalized Environment and Genes Study (PEGS), which includes more than 20,000 North Carolinians.
This study incorporates participants’ health data and environmental exposure information, accounting for factors such as water quality, diet, and air pollution. The goal is to pinpoint drivers of chronic conditions like diabetes and heart disease at an individual level. PEGS is motivated in part by the idea that because people are exposed to multiple substances, research needs to assess the exposome — the totality of a person’s exposures and their biological effects.
Now, Motsinger-Reif is helping to apply that exposomics framework to the National Institutes of Health All of Us Research Program.
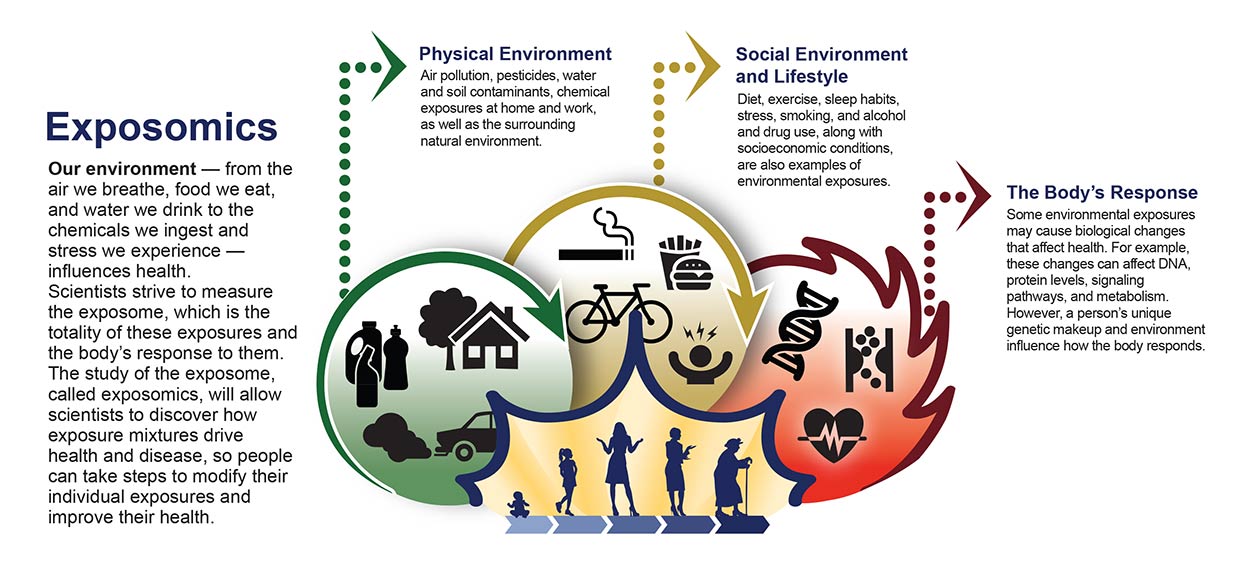
“We recently launched a collaborative effort with All of Us where we are analyzing stored blood samples from 5,600 participants, focusing on those who developed Type 2 Diabetes after joining,” she said. “We are assessing whether a variety of environmental exposures interact with genetic and behavioral factors in a way that correlates with health outcomes.”
Research into gene-environment interactions has been aided by rapid advances in computing power, and that trend will continue, noted Motsinger-Reif during the Q&A session.
“Ten years ago, it would have taken 100,000 human years to compute the data we can now analyze in a fraction of the time,” she said. “This is an exciting shift.”
Studying diet’s role in cancer
Wade shared research from his lab demonstrating how a high-fat diet can spark epigenetic changes that may increase cancer risk. His team fed mice a diet of 10% fat, while another group received 60% fat. After 20 weeks, the high-fat group showed epigenetic markers associated with colon cancer.
“The mice did not develop colon cancer but showed gene expression changes suggesting this could be a likely outcome if they remained on the diet,” Wade explained.
He added that the research raises important questions about the long-term effects of obesity. Even after mice on the high-fat diet lost weight, 25% of the markers of colon cancer still existed.
“Keep in mind that the mice grew to the equivalent of a 400-pound human,” Wade noted. “If we had them grow to a more optimal size, we’d have to see whether the epigenetic changes were as significant.”
An attendee asked Wade how his research might apply to a ketogenic diet, where fat can be included in higher amounts.

“We have studied ketogenic diet in rodents, and although we do see some negative reactions in genes, they are not as significant as what we find in our high-fat model,” said Wade. “This tells me that metabolism plays a huge role in how genes react to diet.”
Wade was asked how he sees epigenetics evolving in the next few years.
“We have genome sequences and the tools to study them,” said Wade. “The only thing holding us back is our imagination. Years from now, people will look back on this time as a golden era in the field.”
(Sam Tyler is a technical writer-editor in the NIEHS Office of Communications and Public Liaison.)