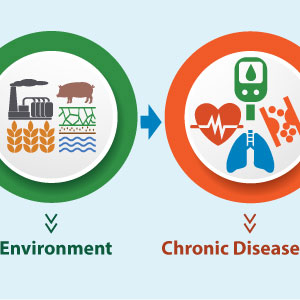
To deliver on the promise of precision medicine, where disease prevention, diagnosis, and treatment are tailored to a patient’s unique biology, we must look beyond just genetics. That’s because our environment — the air we breathe, the food we eat, the stress we experience, and so forth — also plays a key role in shaping health outcomes.
In addition, we know that such exposures do not occur in isolation but rather interact throughout our lives. So, in my view, to advance individualized healthcare, analysis of the exposome — a person’s cumulative exposures and their corresponding biological effects — should go hand in hand with analysis of the genome, the complete set of a person’s DNA.
I recently had the pleasure of speaking with Konstantinos Lazaridis, M.D., executive director of the Mayo Clinic’s Center for Individualized Medicine. He offered great insight into how integrating exposomics with genomics will enhance personalized healthcare approaches. Dr. Lazaridis outlined how a large-scale collaborative effort akin to the Human Genome Project — a Human Exposome Project — could revolutionize our understanding of disease.
We discussed the practical challenges of bringing exposomics into clinical practice, and the role of artificial intelligence in managing and analyzing the vast data involved. Dr. Lazaridis also shared what inspired him to pursue a career as a physician-scientist.
Expanding the scientific toolkit
Rick Woychik: What is precision medicine, and how do you see it evolving in the coming years?
Konstantinos Lazaridis: Precision medicine aims to apply novel approaches to better treat patients by individualizing prognosis, diagnosis, and drug therapy. If we go back 10 or 15 years, we diagnosed and treated everyone the same. But then pharmacogenomics was introduced — identifying the right drug for the right patient at the right time. We learned how each of us metabolizes medications differently. Then we began to focus on identifying the cause of malignancies by analyzing both hereditary mutations and somatic mutations [changes in DNA that occur after conception]. This is what we now call precision medicine.

These days, the term "precision medicine" is more commonly used than "individualized medicine." I tend to prefer the term individualized medicine because precision medicine, to me, has traditionally focused on cancer, but there is so much more to understand and treat beyond cancer. There are inflammatory diseases, aging-related diseases, and environmentally induced diseases, including cancer, that we know very little about today.
A few years ago, at our Center for Individualized Medicine, the main focus was genomics. It still is a major focus, but there's a significant shift in our vocabulary now. We're actively working to incorporate more "omics" and AI to better understand disease because, while genomics is key, there are several other omics and tools we need to explore.
In my view, those other omics — proteomics, transcriptomics, methylomics, microbiomics, and so forth — reflect the broader impact of exposures on our biology, to say nothing of exposomics itself. The ones I mentioned provide critical knowledge about proteins we produce, gene expression patterns, chemical modifications regulating genes, and the role of microorganisms in our health.
I consider all these non-genomic omics as part of the broader exposome, and wherever possible, they should be incorporated into traditional genomics research. Studying the interplay of these different omics and their connection to disease will be groundbreaking. Of course, all of this work will be challenging, but there is tremendous opportunity to better understand these interactions in order to develop novel therapies.
I envision that 10-15 years from now, we won’t just assess individuals for their genomes. We’ll evaluate them based on their methylome, proteome, exposome, and so forth. This comprehensive assessment is needed to understand their overall health and disease prognostication at a more detailed level, so that we can provide an individualized care plan.
Advancing a new paradigm
RW: There’s been a lot of discussion about launching a Human Exposome Project, similar in ambition to the Human Genome Project. What are your thoughts on such an initiative?
KL: I think about this often, as the exposome is the next frontier of medicine in several ways. We need to revisit datasets built around large study populations and take advantage of all of that scientific knowledge at our disposal. As you know, there are important efforts like the NIH All of Us Research Program and the UK Biobank, and several prominent institutions have initiatives around genome sequencing. I think that leveraging those efforts and datasets will be critical. After all, the exposome doesn’t work in a vacuum — it works in interaction with the genome.
At Mayo Clinic, we’ve spent the last five years sequencing the exomes of about 100,000 individuals seen in our practices. The exome refers to all — about 20,000 — genes in the genome that produce proteins. We now have available those individuals’ exome sequences and are providing access of these data to more than 120 scientific groups conducting research in more than fifty diseases. As we move forward, we’re creating a smaller cohort of about 50,000 patients to start doing methylomics, transcriptomics, proteomics, and microbiomics testing using blood, urine, stool, and saliva samples. The goal is not only to analyze each kind of omics data separately but also to integrate several omics together in a way that will be informative and help to better elaborate disease processes.
We are strategic about what we test because we can’t afford to do everything at once. But imagine what we could learn from 50,000 people if we perform, for example, methylome testing to estimate their biological clocks to examine age acceleration, and then correlate those findings with other omics information and robust phenotypic data. That’s the kind of paradigm we want to advance.
EKG, meet AI
RW: That is fascinating, and your center’s work will go a long way toward advancing exposomics and individualized medicine. On a related note, I am excited about a new initiative funded by NIEHS that will expand research into the exposome, which was announced just last month. The Network for Exposomics in the U.S. will bring together leaders in the field to operationalize exposomics across the country and provide a vital complement to genomics.
Bringing to bear cutting-edge approaches, such as the omics you mentioned and tools such as mass spectrometry, will allow researchers to incorporate exposome analysis into a wider breadth of biomedical research, which is positive step forward.
Speaking of cutting-edge tools, I understand that your team at Mayo has been able to successfully incorporate AI into some of your research efforts. Can you expand on that?
KL: Sure. At Mayo, we’ve made significant progress to understand how AI can help us predict future events using electrocardiogram, or EKG, data. Back when I was in medical school, we believed that an EKG, which provides an electrical profile of the heart, only tells you about the past and the present, not the future. Now, we realize that when you use AI, there are EKG patterns the algorithm can recognize that humans cannot. So, for example, using existing EKGs, we can predict the development of atrial fibrillation, congestive heart failure, aging of the heart, and even non-cardiac diseases like fatty liver.
I’d like to go back and not just to look at our patients’ exomes and genomes, but also at other omics analyses to see whether they explain some of the features we observe. For example, we’re finding people who are 50, but the EKG algorithm tells us they’re biologically 55 or even 60. This is an observation I’d like to try to explain. Is this observation due partially to the exposome? Is it methylation-driven? Is it protein-driven? And with such knowledge, can I reverse that or slow the biological aging of the heart? This is where exposures such as diet, lifestyle, and others come into play, and these are the key elements we’d like to explore more.
Again, we need to leverage big cohorts like All of Us, the UK Biobank, and others where we have phenotypic information on many thousands of individuals. We can use that scientific knowledge to say, okay, we know about genomics, which explains X. But now let’s go beyond to see what methylomics can tell us and what proteomics can reveal, as just two examples. If we integrate such analyses properly, we’re going to fill a lot of knowledge gaps about what drives the phenotypes we observe, and specifically the types of environmental exposures at play that impact biology.
Accounting for complexity
RW: Any final thoughts for Environmental Factor readers?
KL: For a long time, we thought hemochromatosis — a condition that leads to chronic liver damage and cancer because the patient can’t stop absorbing iron from the gut — was solely a genetic disease. But for some time now, I would argue that it’s related to the environment as well. If iron did not exist in the environment, you might never develop hemochromatosis, regardless of whether you have a genetic defect. This highlights the importance of the environment and how we can start thinking along these lines.
Through genomics, we've made significant discoveries on many diseases, provided valuable answers to patients’ needs, and changed how we practice medicine. But when it comes to many diseases, particularly common ones, genetics alone helps to explain only a small percentage of those conditions. This means that in most cases, we are concurrently dealing with both genetics and environmental factors. Our research strategy needs to better account for that complexity, and I think exposomics will help us do exactly that.
(Rick Woychik, Ph.D., directs NIEHS and the National Toxicology Program.)