New informatics and biomonitoring technologies could allow researchers and regulators to make the most of data sets for investigating the health impacts of environmental exposures, according to Xiuxia Du, Ph.D., from the University of North Carolina at Charlotte. Du introduced the technologies during her Jan. 30 Keystone Science Lecture.

In work funded by NIEHS and others, Du’s laboratory has developed a set of informatics tools collectively called Automated Data Analysis Pipeline (ADAP). The goal is to better process and integrate an increasing flood of exposomics and metabolomics data in public repositories. Her team also developed a separate set of informatics tools that work with portable biosensors for onsite biomonitoring of exposures.
Building meaning from data
Using ADAP, researchers can explore existing, publicly-available research in metabolomics — a method to identify and quantify small molecules in biological or environmental samples — and exposomics, which refers to measurements of environmental exposures across the lifespan. These and similar approaches to view biology from a global perspective are often called “omics.”

“Dr. Du’s research will provide insight into the integration and harmonization of important omics data across cohorts and studies,” said Yuxia Cui, Ph.D., a program officer in the NIEHS Division of Extramural Research and Training and Du’s host for the Keystone Science Lecture event.
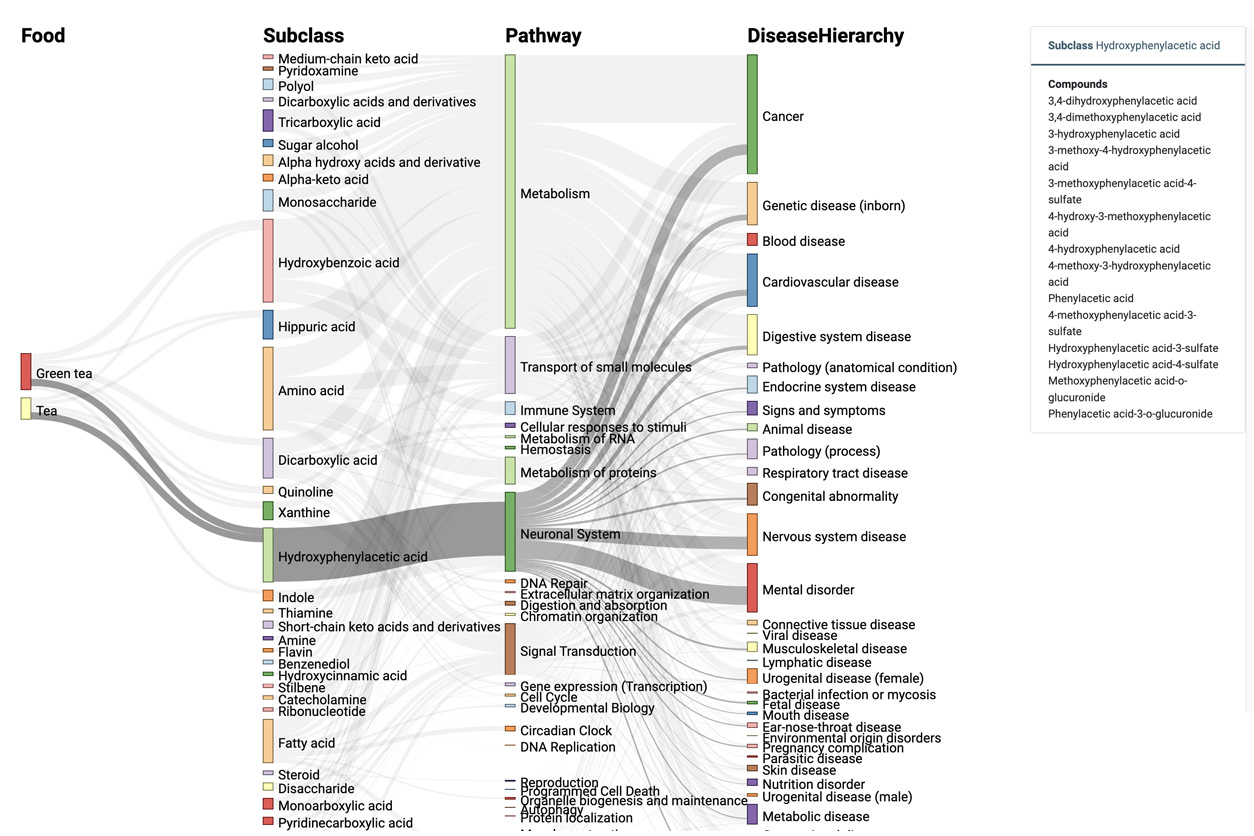
Linking pathways from food to disease
Du and her team created the ADAP-BIG and ADAP-KDB systems, which can analyze, prioritize, and characterize previously unknown environmental compounds. In particular, based on data acquired for different studies by different laboratories, they looked for a consensus among structural themes in their results.
In addition, MetaboFood, a web resource that Du’s laboratory has developed through close collaborations with colleagues Jing Yang, Ph.D., and Colin Kay, Ph.D., will be integrated with the ADAP informatics framework. MetaboFood can help scientists identify new links between food, metabolic pathways, and diseases. Currently, the system has data for 17 types of food, including apples and tea. The technology can display similarities and differences in specific food compound compositions.
“We can look at how foods can affect immune system diseases and check metabolic pathways,” said Du. Further developments are currently underway to incorporate more types of food into MetaboFood.
Benefiting everyone
Researchers can download ADAP-BIG and deploy the system on a single computer or on a high-performance computing cluster. The free technology measures up to paid data analytics programs in many cases, said Du. In comparison tests, the researchers found that ADAP-BIG performed as well as fee-based software tools. ADAP-KDB and MetaboFood are freely available web resources that Du welcomes everyone to test and use.
Du welcomes researchers to upload their own data, especially mass spectra, to ADAP-KDB or other publicly available data repositories. By doing this, the information will be more widely available for others and can benefit the broad metabolomics and exposomics communities, thereby maximizing the value of the data.
Du’s team developed and continues to improve the ADAP informatics in collaboration with researchers at University of North Carolina at Chapel Hill, RTI International, University of Michigan, Colorado State University, Washington University in St. Louis, and University of North Carolina at Charlotte.
Citations: Smirnov A, Liao Y, Fahy E, Subramaniam S, Du X. 2021. ADAP-KDB: A spectral knowledgebase for tracking and prioritizing unknown GC-MS Spectra in the NIH’s metabolomics data repository. Anal Chem 93(36):12213-12220.
Du X, Smirnov A, Pluskal T, Jia W, Sumner S. 2020. Metabolomics data preprocessing using ADAP and MZmine 2. Methods Mol Biol 2104:25-48.
(Catherine Arnold is a contract writer for the NIEHS Office of Communications and Public Liaison.)









