Williams’ winning study represents a full circle in his career. As a doctoral student, he studied how mutations in the BRCA1 protein compromised its functions. (Photo courtesy of Steve McCaw / NIEHS)
Williams’ winning study represents a full circle in his career. As a doctoral student, he studied how mutations in the BRCA1 protein compromised its functions. (Photo courtesy of Steve McCaw / NIEHS)On March 18, Scott Williams, Ph.D., deputy chief of the NIEHS Genome Integrity and Structural Biology Laboratory, received the Southeast Regional Collaborative Access Team (SER-CAT) Outstanding Science Award.
Each year, a review panel selects a paper deemed to have the highest scientific impact that was published by a SER-CAT member. “This award is a real honor,” Williams said. “We have been using the SER-CAT facilities since 2010, and this resource has been critical for our research.
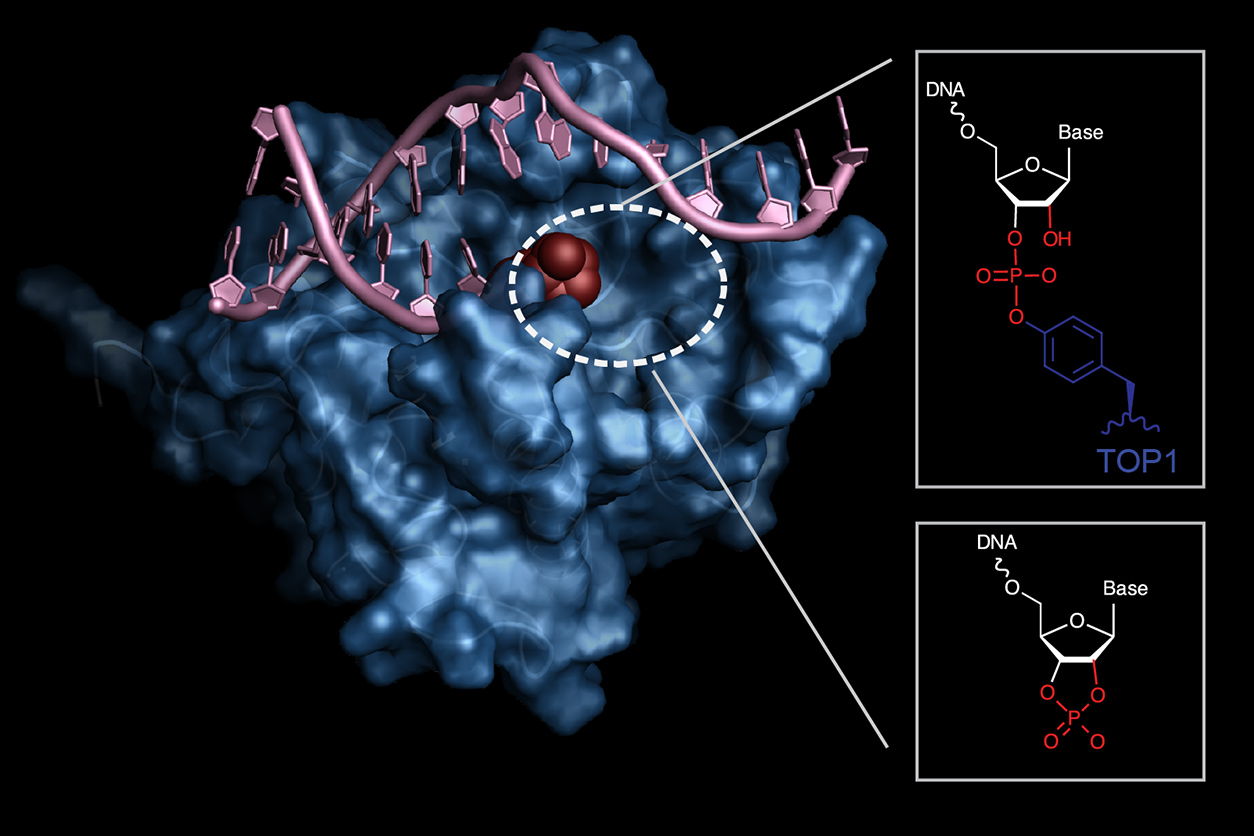
He explained that his lab routinely uses synchrotron radiation to visualize proteins and protein-DNA complexes at the near atomic scale (see sidebar). Synchrotron radiation is a form of electromagnetic energy that is generated when charged particles accelerate in a curved or orbital path.
The SER-CAT organization, with 21 member institutions, was formed in 1997 to provide advanced X-ray capabilities to scientists in the southeastern region of the United States. SER-CAT is located at the Argonne National Laboratory Advanced Photon Source (APS) and is operated by the University of Georgia.
Scoping out molecular structures
Williams aims to understand how DNA repair mechanisms can be used in treatment of diseases like cancer. He uses a technique called macromolecular crystallography to study how the body recognizes when DNA is damaged after environmental exposures, and how it is repaired.
He also studies how mutations affect proteins that protect genome stability, in syndromes that predispose certain people to cancer or neurological disease.
“The SER-CAT Outstanding Science Award for 2021 to Scott Williams is a well-deserved high honor and a testament to his success in solving structures through the SER-CAT,” said Bill Copeland, Ph.D., head of the Genome Integrity and Structural Biology Laboratory. Williams is in good company ― in 2014, his colleague Samuel Wilson, M.D., won the award.
“Williams’ group has published over 50 structures in 20 papers during his time at NIEHS — all of which have benefitted from SER-CAT,” Copeland noted.
 Aerial photo of the APS at Argonne National Laboratory, Argonne, Illinois, USA. (Photo courtesy of Argonne National Laboratory, John Hill / Tigerhill Studio, under Creative Commons license CC BY-NC-SA 2.0)
Aerial photo of the APS at Argonne National Laboratory, Argonne, Illinois, USA. (Photo courtesy of Argonne National Laboratory, John Hill / Tigerhill Studio, under Creative Commons license CC BY-NC-SA 2.0)Breast cancer insights
Williams earned the SER-CAT Outstanding Science Award for his paper “Endogenous DNA 3’ blocks are vulnerabilities for BRCA1 and BRCA2 deficiency and are reversed by the APE2 nuclease,” published last year in the journal Molecular Cell.
The research was part of a multidisciplinary collaboration with Dan Durocher, Ph.D., from the University of Toronto. The team reported that cancer cells with mutated BRCA1 and BRCA2 genes died when they lacked a protein called apurinic endonuclease 2 (APE2). This knowledge could be applied to personalized medicine in the future and potentially improve breast cancer outcomes.
On March 18-19, Williams participated in the St. Jude-SERCAT Structural Biology Symposium, which occurred virtually this year. During his award lecture, Williams discussed his lab’s work with SER-CAT beamlines (see sidebar and diagram below) to determine the molecular structure of the APE2 nuclease, a new promising anti-cancer drug target.
Citations:
Alvarez-Quilon A, Wojtaszek JL, Mathieu M-C, Patel T, Appel CD, Hustedt N, Rossi SE, Wallace BD, Setiaputra D, Adam S, Ohashi Y, Melo H, Cho T, Gervais C, Munoz IM, Grazzini E, Young JTF, Rouse J, Zinda M, Williams RS, Durocher D. 2020. Endogenous DNA 3’ blocks are vulnerabilities for BRCA1 and BRCA2 deficiency and are reversed by the APE2 nuclease. Mol Cell 8;78(6):1152−1165.e8.
Schellenberg MJ, Lieberman JA, Herrero-Ruiz A, Butler LR, Williams JG, Munoz-Cabello AM, Mueller GA, London RE, Cortes-Ledesma F, Williams RS. 2017. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 357(6358):1412–1416.
Tumbale PP, Jurkiw TJ, Schellenberg MJ, Riccio AA, O'Brien PJ, Williams RS. 2019. Two-tiered enforcement of high-fidelity DNA ligation. Nature Commun 10(1):5431.
Tumbale P, Schellenberg MJ, Mueller GA, Fairweather E, Watson M, Little JN, Krahn J1, Waddell I, London RE, Williams RS. 2018. Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease. EMBO J 37(14):e98875.
Williams JS, Tumbale PP, Arana ME, Rana JA, Williams RS, Kunkel TA. 2021. High-fidelity DNA ligation enforces accurate Okazaki fragment maturation during DNA replication. Nat Commun 12:482.
(Kelley Christensen is a contract writer and editor for the NIEHS Office of Communications and Public Liaison.)
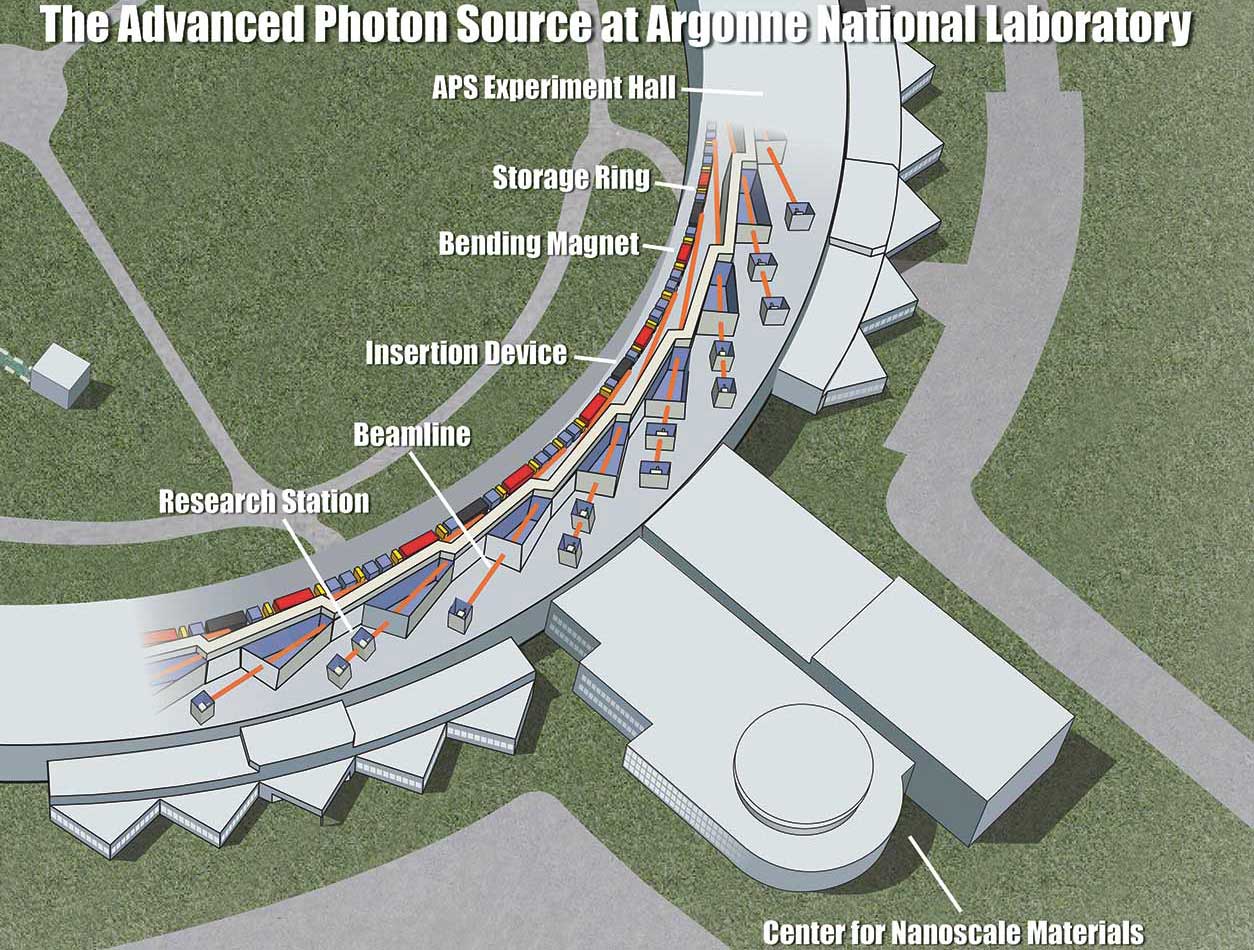
Artist's rendering of the APS with legend. (Image courtesy of Argonne National Laboratory, under Creative Commons license CC BY-NC-SA 2.0)