Patel, shown at a 2018 NIEHS data science workshop, and his postdoc Chirag Lakhani, Ph.D., built an “exposome data warehouse” to support finding geographical, environmental, and social factors linked to health and disease. (Photo courtesy of Steve McCaw / NIEHS)
Patel, shown at a 2018 NIEHS data science workshop, and his postdoc Chirag Lakhani, Ph.D., built an “exposome data warehouse” to support finding geographical, environmental, and social factors linked to health and disease. (Photo courtesy of Steve McCaw / NIEHS)Environmental factors, along with genetics, can play a role in predicting disease. Chirag Patel, Ph.D., from Harvard Medical School, uses data science to analyze the roles of genetics and countless environmental factors in health and disease. He spoke to NIEHS on Dec. 7, as part of the Keystone Science Lecture series.
“Chirag has contributed to the advancement of data-driven technologies for identifying environmental risk factors, as well as gene-environment interactions, for many complex human diseases,” said Kimberly McAllister, Ph.D., a health scientist administrator at NIEHS and host of the talk.
Both genetic factors, or characteristics that are inherited, and environmental factors, such as the circumstance of living with air pollution, contribute to disease development. Scientists have more to learn about these gene-environment interactions.
The general equation
Patel describes his research in the context of a model known as “phenome equals genome plus exposome.”
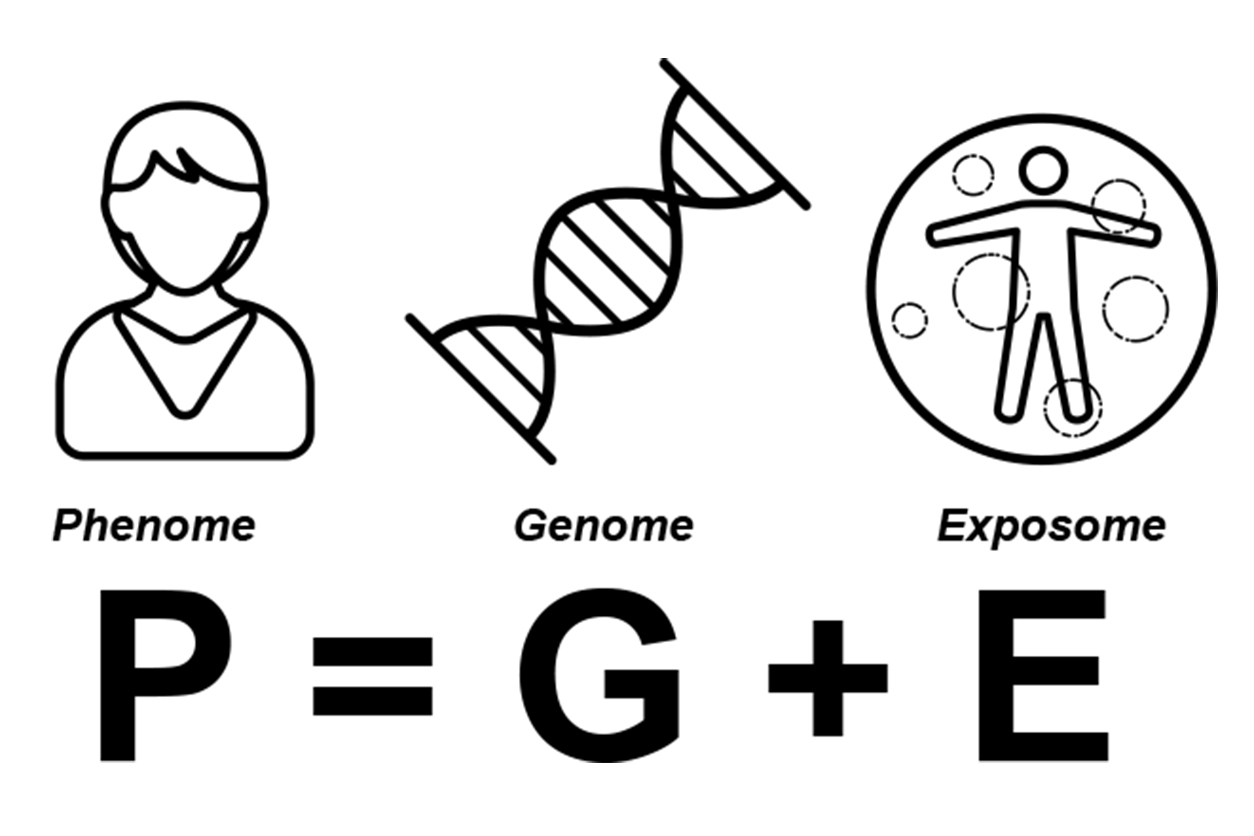 The phenome is the set of phenotypes, the human genome is the complete set of genes, and the exposome is all environmental influences. (Image courtesy of NIEHS)
The phenome is the set of phenotypes, the human genome is the complete set of genes, and the exposome is all environmental influences. (Image courtesy of NIEHS)This general equation captures the notion that the whole being of people, or phenome, results from the interaction of their genes and the environment. A phenome is the set of all phenotypes. Phenotypes are observable characteristics such as height and eye color, as well as health.
The genetic element of the equation, which represents more than 3 billion DNA subunits of the human genome, was decoded in 2003 — in part, by data scientists. During the past decade, an outpouring of human genomic data has fed the growth and accessibility of data sets that allow comprehensive, in-depth studies.
Understanding the second part of the equation — Patel’s frontier — requires comparable quantities of data to study environmental influences. The basis for this line of inquiry is called the exposome, and it represents all of a person's exposures, from before birth to death.
From thousands to millions
According to Patel, one challenge with current environmental health research is that there are not enough human samples available. Studies with thousands of people are considered large. However, to differentiate health differences due to environmental influences, sample sizes in the millions are needed.
Linking data from diverse national or international data sets is essential, and the technologies are at hand.
Patel’s research group works toward that end by developing bioinformatics approaches. His quest to employ existing datasets is a gateway to exploring how genetics and environmental factors influence our health.
 “During this Keystone lecture, we enjoyed hearing about Chirag’s many projects over the past few years,” said McAllister. (Photo courtesy of Steve McCaw / NIEHS)
“During this Keystone lecture, we enjoyed hearing about Chirag’s many projects over the past few years,” said McAllister. (Photo courtesy of Steve McCaw / NIEHS)Still, the machine-level data analysis has confounding variables, making it difficult to determine if specific characteristics of the exposome affect the phenome.
Needles in a haystack
“Our exposome consists of thousands of needles in a huge haystack of disease,” said Patel. “Finding these needles is a massive challenge for data scientists, epidemiologists, and environmental health researchers.”
“Effectively harnessing the diversity of scale and measures of exposures, from the geographical to the molecular, will help us figure out connections,” he continued.
By incorporating machine learning and real-world environmental data, Patel aims to improve overall data quality, enhance discovery of environmental associations with health, and advance predictive capability for complex exposome-phenome relationships.
Next steps
Considering mixtures of chemicals or multiples of exposures at the same time is essential in the next stages of research. It will be accomplished through powerful, innovative data science techniques conducted by interdisciplinary teams.
The resulting characterization of the components of the exposome is a critical action item for boosting precision medicine.
However, an immediate data science question is how to make multiscale and dimensional information useful for both research and the clinic.
Citations:
Rasooly D, Ioannidis JPA, Khoury MJ, Patel CJ. 2019. Family history-wide association study to identify clinical and environmental risk factors for common chronic diseases. Am J Epidemiol 188(8):1563–1568.
Manrai AK, Ioannidis JPA, Patel CJ. 2019. Signals among signals: prioritizing nongenetic associations in massive data sets. Am J Epidemiol 188(5):846–850.
Lakhani CM, Tierney BT, Manrai AK, Yang J, Visscher PM, Patel CJ. 2019. Author Correction: Repurposing large health insurance claims data to estimate genetic and environmental contributions in 560 phenotypes. Nat Genet 51(4):764–765.
Manrai AK, Patel CJ, Ioannidis JPA. 2018. In the era of precision medicine and big data, who is normal? JAMA 319(19):1981–1982.
(Carol Kelly is managing editor for the NIEHS Office of Communications and Public Liaison.)