“Metabolomics is the end product readout of the systems that power a cell,” said Fessler. (Photo courtesy of Steve McCaw / NIEHS)
“Metabolomics is the end product readout of the systems that power a cell,” said Fessler. (Photo courtesy of Steve McCaw / NIEHS)The National Institutes of Health (NIH), which is made up of 27 Institutes and Centers (ICs), is the country’s premiere medical research agency that makes discoveries to improve human health. Each IC has a particular health emphasis, and many of them have facilities called cores that provide their researchers with scientific support. For example, several ICs have cores that sequence DNA or offer X-ray crystallography to study protein structure.
Thanks to a grant written by NIEHS researcher Michael Fessler, M.D., nine ICs with laboratories that use metabolomics — a method to identify and quantify small molecules in biological or environmental samples — have joined together to form the NIH Metabolomics Consortium. Since Fessler is the Principal Investigator, or lead scientist, for funding the consortium, NIEHS became the hub that coordinates research projects for the group (see first sidebar).
'The proposal was to bring these metabolomics labs from across NIH together to standardize procedures that make the work more efficient and to potentially create a cross-cutting core facility that serves all of NIH,' said Fessler.Fessler said that the NIEHS Metabolomics Core Facility (MCF), which is directed by Alan Jarmusch, Ph.D., is playing a central role in the work. Jarmusch and his counterpart at another IC are already working on consortium projects.
 Jarmusch said his expertise is in mass spectrometry, but metabolomics and data analysis have become a primary focus in the last five years. He joined NIEHS in September 2020. (Photo courtesy of Alan Jarmusch)
Jarmusch said his expertise is in mass spectrometry, but metabolomics and data analysis have become a primary focus in the last five years. He joined NIEHS in September 2020. (Photo courtesy of Alan Jarmusch)Snapshot in time
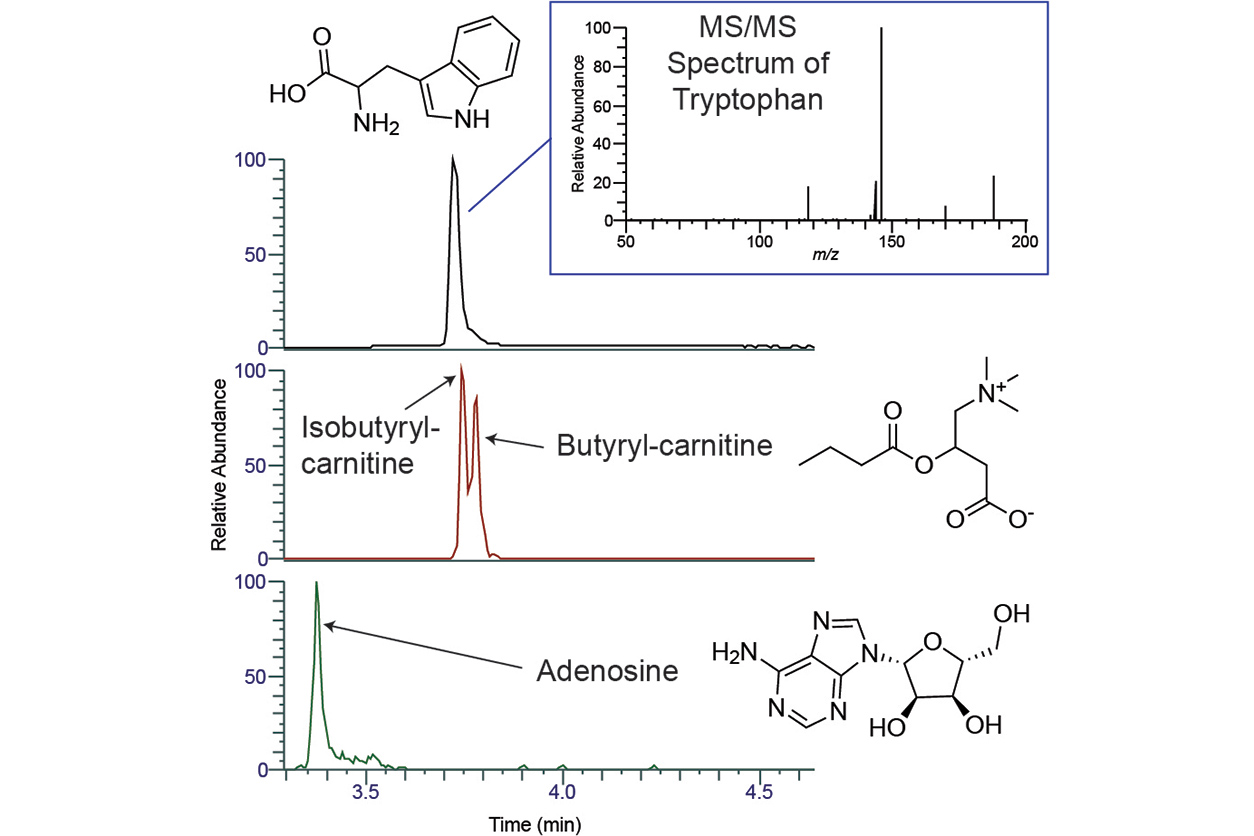
Jarmusch explained that the small molecules he studies, also known as metabolites, represent a snapshot in time of a biological or environmental sample. Using a human cell as an example, metabolomics is the measurement of all the processes in which genes and the proteins they encode are involved. Being able to comprehensively analyze every chemical in a biological sample helps scientists understand the genetic and protein changes associated with disease.
To measure these small molecules, Jarmusch and his colleague, NIEHS chemist Kirsten Overdahl, Ph.D., perform a two-step technique, characterized by chemical separation and mass spectrometry. Chemical separation is primarily performed with liquid chromatography, which separates molecules in a sample based on their polarity, or electrical charge, and similar molecular properties. Mass spectrometry measures a molecule’s mass, which furnishes information on its molecular formula and structure.
Targeted versus untargeted
Fessler and Jarmusch said targeted metabolomics focuses on identifying a limited number of molecules — for example, 10, 20, or 50 — while untargeted metabolomics aims to measure the complete universe of metabolites in a sample. Jarmusch uses the untargeted approach and gave an analogy to illustrate what he does at the MCF.
 Overdahl is excited about the new discoveries NIH researchers will make because of their partnership with the NIH Metabolomics Consortium. (Photo courtesy of Amber Robinson Photography)
Overdahl is excited about the new discoveries NIH researchers will make because of their partnership with the NIH Metabolomics Consortium. (Photo courtesy of Amber Robinson Photography)'If a physician wants to measure the level of vitamin D in a patient, he or she will send a blood sample to the lab that uses the same liquid chromatography and mass spectrometry techniques we use,' said Jarmusch. 'Our approach is not focusing on vitamin D alone but measuring all the chemicals in that blood sample at the same time to gain knowledge about the patient’s physiology.'
Linking to biological knowledge
Fessler and Jarmusch said the NIH Metabolomics Consortium is helping to propel NIH research to new levels of complexity that will benefit mankind, but they want the public to know that data acquisition, or the measurements they take, is one part of the goal. Data analysis, or deriving biological meaning from the measurements, is a huge component of the field of metabolomics that is growing.