The latest Keystone Science Lecture Series featured Yu-Ying He, Ph.D., professor of medicine at the University of Chicago and an expert in a relatively new field known as epitranscriptomics. Also known as RNA epigenetics or RNA modification, epitranscriptomics is the study of the regulation and function of post-transcriptional modifications, the chemical alterations of an RNA molecule. RNA modifications have a crucial role in many human illnesses, such as cancer, obesity, and diabetes, and they influence aging.
 He is also Associate Director for Shared Resources in the Comprehensive Cancer Center at the University of Chicago. (Photo courtesy of Yu-Ying He)
He is also Associate Director for Shared Resources in the Comprehensive Cancer Center at the University of Chicago. (Photo courtesy of Yu-Ying He)On July 20, He presented 'RNA Methylation in Stress Response and Environmental Carcinogenesis' virtually. Frederick Tyson, Ph.D., and Michael Humble, Ph.D., both Health Scientist Administrators in the Genes, Environment, and Health Branch of the NIEHS Extramural Research and Training Division, co-hosted the seminar (see sidebar).
Gene-environment interactions
At the beginning of her presentation, He provided a brief overview of epitranscriptomics and environmental carcinogenesis, or how compounds in the environment contribute to cancer. Environmental compounds such as arsenic inhibit autophagy, or an organism’s ability to remove damaged biomolecules. Some research suggests autophagy is one of the processes that when curtailed can lead to tumor formation.
According to He, there are more than 100 RNA modifications that latch onto RNA, looking like a strand of holiday lights. The modifications regulate all kinds of RNA, such as messenger RNA and transfer RNA, and can regulate RNA decay.
One of the most common RNA modifications is called N6-methyladenosine (m6A), which adds a methyl group to an RNA strand. Other scientists determined that a protein called FTO demethylates or removes the m6A modification.
He’s research found that m6A RNA methylation happens during the stress response and is a critical part of the gene-environment interaction. For many years, though, scientists did not know how m6A worked.
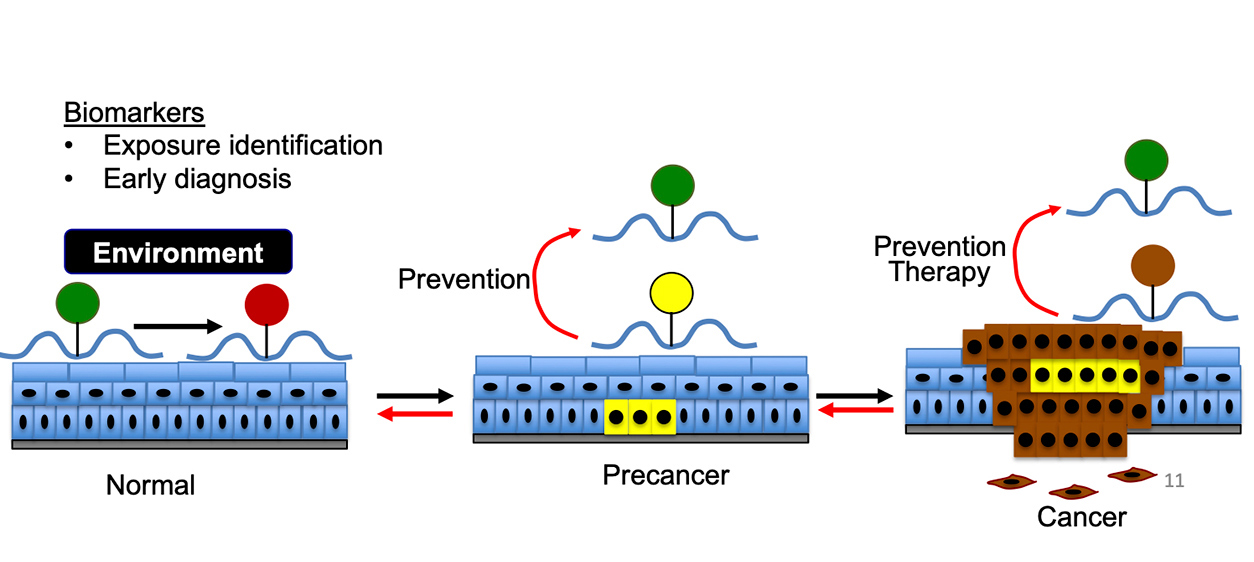
The role of RNA modifications can be seen in tumor development. Normal modifications (green circles) can change into abnormal modifications (red circle) in normal skin, and abnormal RNA modifications (yellow and brown circles) can become normal following either prevention efforts or therapy. (Illustration courtesy of Yu-Ying He)
'Although m6A was discovered in the 1970s, the field didn’t progress much because the enzymes that control m6A were unknown until recently,' He said. 'In 2012, the development of m6A sequencing technology drove the field forward rapidly.'
RNA modifications in skin cancer
A research group led by He studies the early stages of carcinogenesis when cells or mice are exposed to environmental toxins. They concentrate on skin cancer because it is easy to detect such cancer early. He said approximately 3.5 million cases of skin cancer are diagnosed in the U.S. each year, with the main cause being ultraviolet radiation. However, arsenic in drinking water can also be a strong risk factor for developing skin cancer.
Using genetic mouse models, He and her team can manipulate a particular RNA modification enzyme in the skin and focus on the mechanisms that drive the formation of early cancer lesions. They use m6A sequencing to map RNA modifications.
He found that arsenic interacts with an autophagy receptor called p62, which leads to the degradation of the FTO protein. When there is too much FTO, m6A numbers decrease, compromising the stability of the RNA strand, leading to cancer.
'This is an exciting time for research in environmental epitranscriptomics,' Tyson said. 'Dr. He’s high-impact contributions are part of a growing body of evidence identifying mechanistic relationships between exposures, perturbations of epitranscriptomic processes, and disease pathogenesis.'