New tools for analyzing single cells and single molecules hold great potential for the field of environmental health, according to experts who gathered March 7-8 for an NIEHS-sponsored meeting. The tools are advancing at breakneck speed, but their demands on computing power and potential confounding elements call for caution in applications such as risk analysis.
 Kaminski served for three years on the NIEHS National Advisory Environmental Health Sciences Council. He is shown here at a 2017 meeting of the council. (Photo courtesy of Steve McCaw)
Kaminski served for three years on the NIEHS National Advisory Environmental Health Sciences Council. He is shown here at a 2017 meeting of the council. (Photo courtesy of Steve McCaw)These insights were among the outcomes of the latest workshop from the National Academies of Sciences, Engineering, and Medicine (NASEM) Committee on Emerging Science for Environmental Health Decisions, held in Washington, D.C. at the National Academies of Sciences building.
Organizing committee chair Norbert Kaminski, Ph.D., from Michigan State University (MSU), observed that the work reaches beyond biology into the fields of mathematics, engineering, and physics. With abilities such as quantifying the presence of a protein, or mapping biological responses to exposure, the technologies could revolutionize environmental health research.
“These workshops really make a difference in moving us into these [new] areas,” said Linda Birnbaum, Ph.D., NIEHS and National Toxicology Program director, as she welcomed participants. The speakers and panelists who spoke during the workshop outlined the capabilities of the new tools and described early applications in environmental health research.
Unprecedented precision
“We are making measurements using these new approaches … at a resolution and precision that are really unprecedented,” Kaminski observed. “[This] shows the importance of this convergence of different disciplines … if we really want to address some of the difficult questions.”
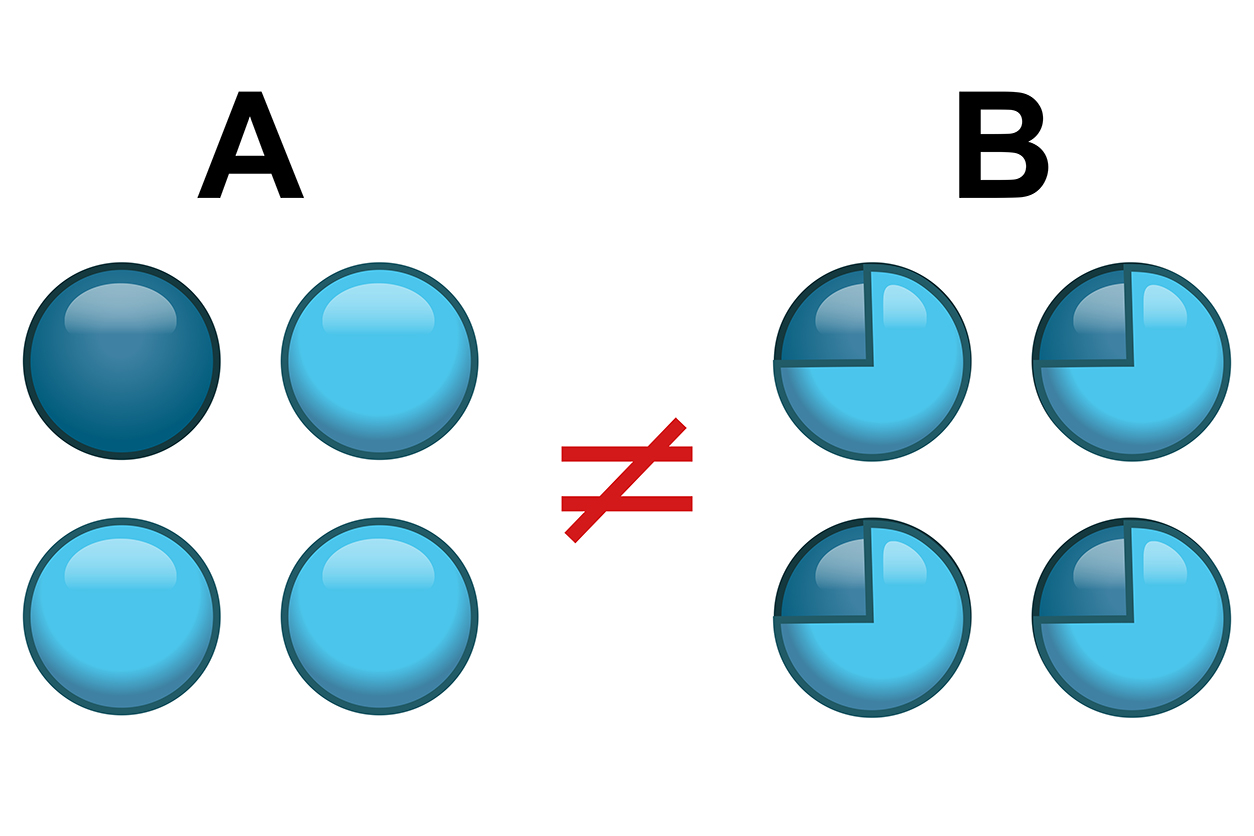 Single cell analyses avoid errors introduced by conventional methods that process hundreds of thousands of cells at a time and average out the differences, Kaminski explained. Speakers reviewed techniques that distinguish individual cells. (Photo courtesy of NIEHS)
Single cell analyses avoid errors introduced by conventional methods that process hundreds of thousands of cells at a time and average out the differences, Kaminski explained. Speakers reviewed techniques that distinguish individual cells. (Photo courtesy of NIEHS)Many speakers underscored the tremendous promise these tools hold for environmental health researchers. “A prime example would be to facilitate better biomarkers of effect,” said Kimberly Thigpen Tart, J.D., an NIEHS health science policy analyst.
Julie Biteen, Ph.D., from the University of Michigan, tracks the movement of single molecules to observe processes in cell biology. In one example, she said her team can pinpoint a molecule’s position to within nanometers.
Bioinformatician Lana Garmire, Ph.D., from the University of Michigan Medical School, creates tools such as Granatum to guide genomics researchers who sequence RNA from a single cell to study its response to a stimulus, such as a chemical exposure. Garmire characterized single cell analysis as an accelerating train. “And computation … is a limiting factor,” she said.
Heavy demand on computing
 Garmire is prototyping a web server to serve as a matchmaker between scientists with data and bioinformatics analysis. “It's like consumer-based Airbnb but in the single cell analysis domain,” she quipped. (Photo courtesy of NASEM)
Garmire is prototyping a web server to serve as a matchmaker between scientists with data and bioinformatics analysis. “It's like consumer-based Airbnb but in the single cell analysis domain,” she quipped. (Photo courtesy of NASEM)The enormous data sets created by single cell and single molecule analysis place heavy demands on computing resources and budgets. Participants discussed concerns about equity, reproducibility, and accessibility for early career scientists and those at smaller institutions.
Potential solutions included organizational and administrative changes, and commercialization. “There is a lot of room for hope,” said Biteen.
Cell type, cell state
The nature of the cell and the timing of the sample also present challenges. For example, cancer tumors may be seen as complex ecosystems, according to Orit Rozenblatt-Rosen, Ph.D., from the Broad Institute. She works to understand molecular-level processes within those ecosystems to find the best treatment combinations for individual patients.
The variety of cell types within a tissue or organ can complicate how scientists studying the origins of disease determine which tissues, cells, or genes are being disrupted. Mike Mancini, Ph.D., from Baylor College of Medicine, studies endocrine-disrupting chemicals such as bisphenol A. For example, he observed that cells from a single tumor vary in the number and distribution of their hormone receptors.
Rajanikanth Vadigepalli, Ph.D., from Thomas Jefferson University, underscored what Birnbaum referred to as daunting complexity. From a control systems viewpoint, he suggested, “There is no normal [cell] state, rather [there is] a normal trajectory.”
Cells move through states such as developmental transitions, and adaptations and adverse responses to stressors. Consideration of cell state calls for a convergence of single cell assays, bioinformatics, and dynamic modeling and simulation, Vadigepalli explained.
“It becomes increasingly apparent that we need to formulate our questions and problem statements related to environmental health decision-making as clearly and practically as possible,” noted organizing committee member Lesa Aylward, Ph.D., of Summit Toxicology. “Otherwise, we will be overwhelmed by detail and will not gain relevant insights into the problems we are trying to solve.”
Citations:
Rozenblatt-Rosen O, Stubbington MJT, Regev A, Teichmann SA. 2017. The human cell atlas: from vision to reality. Nature 550(7677):451−453.
Zhang B, Huang K, Zhu L, Xu W. 2017. Precision toxicology based on single cell sequencing: an evolving trend in toxicological evaluations and mechanism exploration. Arch Toxicol 91(7):2539−2549.