 At NIEHS, Rodriguez will study the seemingly random distribution of RNA expression, including its deep repressive state. (Photo courtesy of Steve McCaw)
At NIEHS, Rodriguez will study the seemingly random distribution of RNA expression, including its deep repressive state. (Photo courtesy of Steve McCaw)Scientists often describe gene expression as an either/or proposition — genes are either on and active, or they are off, and thus, inactive. However, new research indicates that active genes likely spend most of their time in an inactive phase, called the deep repressive state. The findings, published Dec. 13 in the journal Cell, may lend insight into how variability in gene expression can contribute to disease.
“Our study is one of the real firsts in observing how human genes respond to hormones in real time,” said lead study author Joseph Rodriguez, Ph.D., a new faculty member in the NIEHS Epigenetics and Stem Cell Biology Laboratory (ESCBL). “The field hasn’t really considered that there could be really long periods of inactivity, so [these findings] add a new layer of regulation to the current model of transcription.”
Bring the noise
In the central dogma of molecular biology, genes are transcribed into RNA messages, which are then translated into protein products. That first step — transcription — has been likened to a light switch. Genes that get flipped on are actively transcribed, or expressed. Those that are flipped off are not. Active genes are then switched on and off over time.
Over the last decade, researchers have noticed a lot of variability in the way genes are turned on or off, even in identical cells in identical environments. One cell might be producing a couple dozen copies of a particular RNA message at the same time that another cell — in that same tissue — is churning out hundreds.
Rodriguez says this gene expression noise, as he and others call it, gets worse with age and is a hallmark of cancer. “This noise exists in normal tissues, but in cancer, it appears to be hijacked to allow even more diversity in the tumor population, making it easier for tumor cells to develop resistance to treatment,” said Rodriguez. “Understanding the origins of this variability, and how gene expression is fine-tuned, is one of my goals.”
CRISPR breakthrough
In the new study, Rodriguez focused on breast cancer cells and a gene called TFF1, which is remarkably responsive to estrogen. When scientists expose cells to a form of estrogen called estradiol, the expression of TFF1 can increase a hundred-fold. Rodriguez wanted to watch the process in individual cells, in real time.
To do so, he needed to attach fluorescent tags to the TFF1 gene. He tried for years, with no luck. Finally, the new gene editing technique known as CRISPR allowed Rodriguez to tag multiple copies, or alleles, of the gene in the same cell and record their activity as they turned on and off in response to estradiol.
“We observed some really cool phenomena,” said Rodriguez. “We saw that some alleles were stuck in a long, deep repressive state that could last several days, even when other alleles nearby were flipping on and off. Human transcription is dynamic, more dynamic than we ever realized.”
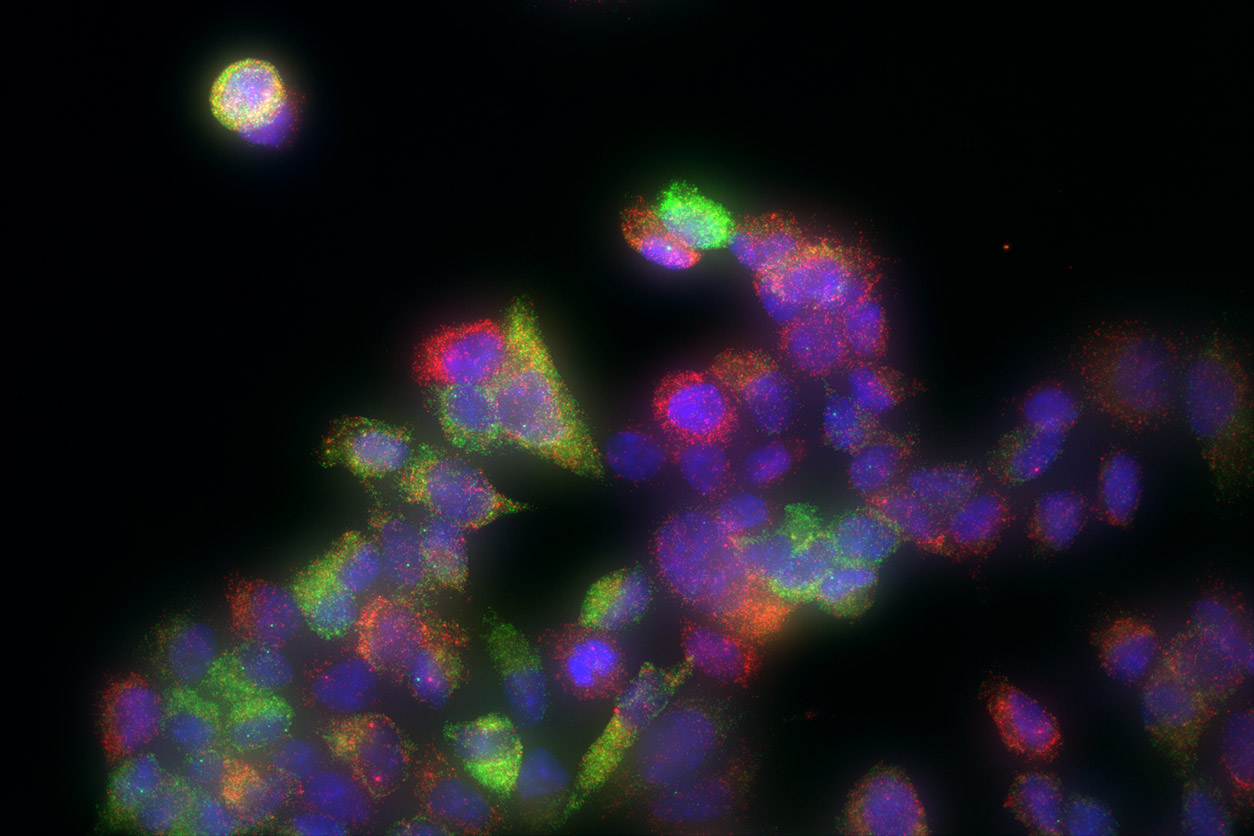 Imaging of individual breast cancer tumor cells shows two different estrogen-responsive genes, one highlighted in green and the other in red. The genes display varying levels of activity in different cells, as shown by the different expressions of color. (Photo courtesy of Joseph Rodriguez, Carson Chow, and Daniel Larson)
Imaging of individual breast cancer tumor cells shows two different estrogen-responsive genes, one highlighted in green and the other in red. The genes display varying levels of activity in different cells, as shown by the different expressions of color. (Photo courtesy of Joseph Rodriguez, Carson Chow, and Daniel Larson)The question of endocrine disruptors
Rodriguez, who led the research as a postdoctoral fellow in the lab of Dan Larson, Ph.D., at the National Cancer Institute, has more questions about this deep repressive state that he hopes to answer in his new post at NIEHS. “Why does this happen? How does it change as a result of environmental exposure? How does it become dysregulated as we age?” he asked.
“Joe brings to ESCBL an entirely novel and innovative approach to evaluating the transcriptional response,” said ESCBL Chief Trevor Archer, Ph.D., adding that the new approach is an excellent complement to strategies other members of the laboratory are using. “I am excited to see the impact that his observations, outlined in this paper, will have on the field of transcription and gene regulation.”
Using his live imaging technique, Rodriguez wants to look at how endocrine disruptors, which can mimic hormones like estrogen, affect expression of genes like TFF1. “I would like to get into the nuts and bolts of how endocrine-disrupting chemicals like BPA [bisphenol A] are functioning and how they impact transcriptional memory,” he said.
Citation: Rodriguez J, Ren G, Day CR, Zhao K, Chow CC, Larson DR. 2018. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. doi: https://doi.org/10.1016/j.cell.2018.11.026 [Online 13 December 2018].
(Marla Broadfoot, Ph.D., is a contract writer for the NIEHS Office of Communications and Public Liaison.)










