Cutting-edge technologies and scientific approaches that promise to complement the use of experimental mammalian models in research have shown great progress in recent years, thanks in part to efforts by leaders at NIEHS and others in the environmental health sciences. Referred to as new approach methodologies (NAMs), the tools range from machine learning and in vitro [cell-based] systems to high-volume chemical screens and sophisticated computational techniques that help scientists integrate and interpret large amounts of toxicological data.
One kind of NAM that I believe could help to transform our ability to understand environmental exposures and their potential biological effects is actually a tiny nonmammalian species — the zebrafish. In early January, I took part in an environmental health conference in India hosted by the country’s National Environmental Engineering Research Institute, which is part of the Council of Scientific and Industrial Research there. Among other presentations, NIEHS grant recipient Robyn Tanguay, Ph.D., from Oregon State University (OSU), discussed her use of zebrafish and why she thinks they represent a paradigm shift in toxicological research.
After the conference, I caught up with Dr. Tanguay — who also directs the NIEHS-supported Superfund Research Center at OSU — to learn more about her scientific portfolio and better understand what makes zebrafish a potentially game-changing model organism. We discussed how zebrafish can help researchers study many chemicals at once, assess the effects of gene-environment interactions, prioritize substances to study in traditional animal models, and shed light on critical biological mechanisms that can be altered due to exposures. I also asked Dr. Tanguay about what inspired her to become a scientist.
Research benefits of zebrafish
Rick Woychik: Why are zebrafish an ideal model to study the potential health effects of environmental exposures?

Robyn Tanguay: The genetic similarities between zebrafish and humans are remarkable because the genes necessary to make a human are quite like the ones necessary to make a fish. By leveraging that similarity and other attributes of zebrafish, researchers can incorporate this model organism in important ways, including to more rapidly identify exposures that are consequential in terms of how they interact with our genetic and biological makeup.
Zebrafish are native to India, and they have different lateral stripes that resemble a zebra. The adults are approximately one inch in length, but we actually do the bulk of our work in the embryonic development stage, where we are then talking about millimeters in length. In that early-life phase, they are transparent, so you can actually observe the development of the brain, heart, eye, ear, and so forth with a simple, inexpensive microscope. Among other benefits, that makes it easy to see how an exposure may affect development of a given organ, for example.
Another major benefit of studying zebrafish is how fast they develop. We can start one morning with a single cell, and by noon the next day, there is already a beating heart, and many of the other organ systems form after just two days. This allows for much greater speed and efficiency in our studies than is possible in traditional rodent systems. The fast development of zebrafish — combined with different imaging capabilities and tools for genetic analysis at our disposal — allows for exciting discoveries to be made at scale and at low cost.
Importantly, almost every gene encoded in an organism is called upon to accomplish a task or multiple tasks in early development. Sometimes, a gene is only transiently needed to accomplish a developmental task, and then maybe later in life it plays an important role in the adult liver, for example. But we know that almost all genes are expressed and active during embryonic development. So, that is an ideal time to see whether a chemical perturbs how genes function and causes observable physical changes that can be associated with a disease outcome.
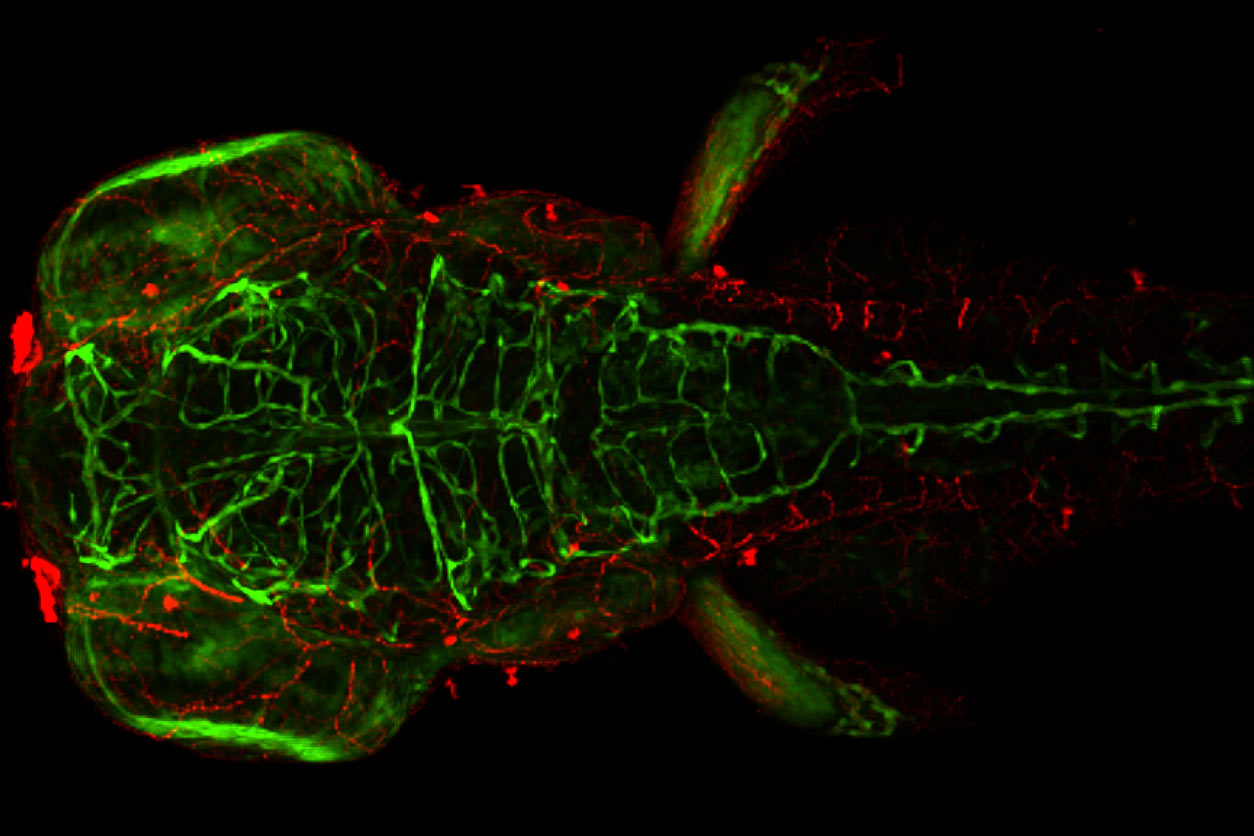
Breakthrough in neurobehavioral assessment
RW: Throughout the years, your laboratory has used zebrafish to study potential health effects of chemicals such as polycyclic aromatic hydrocarbons, flame retardants, pesticides, and complex mixtures. Can you expand on that work for our readers?
RT: Sure. We first sought to observe the normal course of zebrafish development to understand their baseline physical features and behavioral attributes. This could include heart rate, diameter of the eyes, curvature of the body, and so forth. Next, we started doing exposures using individual chemicals from different classes, such as dioxins, metals, and others that you mentioned. The goal was to identify all of the abnormal phenotypes that resulted from exposures, and then to dive deeper by analyzing what genetic and biological mechanisms were involved.
One thing we noticed right away is that there were common physical and behavioral effects caused by many different chemicals, such as heart edema [swelling caused by buildup of fluid]. On its own, that information did not give us great insight into the potential mechanisms involved, but it did show the value of our model to screen for physical and behavioral changes caused by a variety of exposures, which would help to prioritize our follow-up assessments.

Also, we discovered that we could use zebrafish to study neurobehavioral effects at scale, which had not previously been possible and is quite challenging in cell-based models. That meant we could quickly assess alterations in nervous system development, changes in motor activity, and more. Then we sought to do integrated assessments whereby we measured all phenotypes in a matter of days, at numerous concentrations. A lot of my group’s early innovation in this area came through NIEHS funding, when we were challenged to do something that could represent a paradigm shift for the field. I think our work had that effect, and I am grateful for that support.
Tracing cause and effect
RW: How has your team sought to move from identifying a phenotype to better understanding what genetic and biological mechanisms may be causing it? And how can such knowledge be translated to human contexts?
RT: In the field of toxicological risk assessment, there is increased interest in identifying gene expression changes that are predictive of a disease outcome and then determining what exposure level is associated with that outcome. That is the same kind of scientific knowledge we seek to gain through our work with zebrafish. We use transcriptomics [large-scale assessment of gene expression changes in an organism] to identify what genetic changes may result from lower exposure levels that are more like what humans can experience in everyday life.

When we identify those gene expression changes, we can try to see what changes are adaptive or not. As humans, we get exposed all of the time to different substances, but our bodies respond adaptively to change metabolism or eliminate a chemical, and then homeostasis is restored. It is when you hit those tipping points — where you either disrupt too many genes or modulate the activity of a genetic process and alter homeostasis — where disease kicks in.
By looking at exposures that both affect gene expression and produce phenotypes, we hope to identify a causal relationship — one showing that an exposure not only caused a phenotype but also that the phenotype resulted due to a specific genetic mechanism. Gene-editing technologies at our disposal allow us to robustly test what aspects of the genetic landscape may play a role in a phenotype that we observe. Also, because of the small size and transparency of zebrafish, we can look in real time at how cells interact with each other after an exposure and change in gene expression, which further aids our analysis.
When it comes to using knowledge from zebrafish studies in decision-making contexts, I know that regulators are now receiving zebrafish data as part of their informational packets. I have worked closely with the EPA [U.S. Environmental Protection Agency] and the FDA [U.S. Food and Drug Administration] to help them become more comfortable with this new type of data stream. I view data regarding a particular exposure in a zebrafish model as a case study whereby we can identify correlations with traditional research data and, more importantly, try to understand why there may be divergence, which could help to strengthen the use of this model. I am confident that going forward, zebrafish data will be used by decisionmakers to help make more informed choices that can impact public health for the better.
Shedding light on PFAS
RW: Speaking of human-relevant exposures, I understand that your laboratory has started looking at one that has grabbed news headlines in recent years — per- and polyfluoroalkyl substances, or PFAS. Can you share a little bit of your initial work in this area?
RT: The diversity of these structures is remarkable. We have screened hundreds of them in our zebrafish system, and I can say that they absolutely do not behave the same way. Some can bioaccumulate, but others do not, and some are metabolized while others are active at low concentrations. So, we are trying to use our model to better understand the features of those chemicals that make them more or less hazardous.
We want to identify PFAS that bioaccumulate and have specific biological targets, which are the scariest kinds of molecules. Substances that bioaccumulate but do not have specific targets could be a problem with long-term exposures, but I am not as worried about those as I am about the ones that hit a biological target and modulate it. We hope that our zebrafish system will help to prioritize which PFAS might be safe for use and which ones should be eliminated.
[To learn more about Tanguay’s zebrafish research and recent publications, please visit her laboratory's website.]
(Rick Woychik, Ph.D., directs NIEHS and the National Toxicology Program.)