Though DNA is typically portrayed as a double helix, its long strands spend much of their time coiled, condensed, and crammed into a tight little package called chromatin. Now, a new study by researchers at NIEHS and the University of Tokyo shows how a specialized protein called GATA3 unfurls chromatin so other proteins can enter and turn on genes.
 Before joining NIEHS in 2004, Wade was an assistant professor in the Emory University School of Medicine Department of Pathology and Laboratory Medicine. (Photo courtesy of Steve McCaw)
Before joining NIEHS in 2004, Wade was an assistant professor in the Emory University School of Medicine Department of Pathology and Laboratory Medicine. (Photo courtesy of Steve McCaw)The study was published August 18 in Nature Communications.
Understanding how GATA3 works normally could give insight into the diseases that arise when it does not work properly. For example, GATA3 is one of three genes mutated in more than 10% of breast cancers.
“This process is absolutely critical to normal biology, development, and response to the environment,” said co-senior study author Paul Wade, Ph.D., deputy chief of the NIEHS Epigenetics and Stem Cell Biology Laboratory.
Protein pioneers
Cells can sense and respond to a wide variety of environmental cues, such as a flash of heat, a flood of chemicals, or a lack of oxygen. One of the first things cells do to adapt to the outside world is to selectively activate or transcribe the genes that provide blueprints for making proteins they need. Proteins called transcription factors control this process by binding to a specific DNA sequence and flipping on other genes.
Many transcription factors cannot bind target sequences that are wrapped up in chromatin. However, a special subset of transcription factors called pioneer transcription factors can breach the chromatin barrier, find their binding site, and open up the structure to other transcription factors. “Pioneer means they're the first ones there. They break the soil to allow the downstream events to occur,” noted Wade.
Geometry matters
Wade has spent years studying GATA3, a pioneer transcription factor associated with the progression of breast cancer. In January, he traveled to Tokyo to spend a sabbatical working in the laboratory of Hitoshi Kurumizaka, Ph.D., whom Wade has known since they were postdoctoral fellows together at the National Institutes of Health in the 1990s.
 Wade and Kurumizaka, back row, fourth and third from the right, respectively, posed with members of Kurumizaka’s laboratory after dining out in Tokyo. (Photo courtesy of Paul Wade)
Wade and Kurumizaka, back row, fourth and third from the right, respectively, posed with members of Kurumizaka’s laboratory after dining out in Tokyo. (Photo courtesy of Paul Wade)Specifically, the researchers wanted to know whether the position of the GATA3 binding sites within chromatin affected the transcription factor’s ability to form an enhancer site that activates nearby genes. “It’s like creating a light switch from a blank wall,” said Wade. “This paper was specifically asking questions about how the local architecture of DNA might determine whether the light switch gets created or not.”
Wade and his collaborators conducted a series of biochemistry experiments in which they moved the GATA3 binding site to different locations and then assessed the outcome. They found that changing the spacing and orientation of the binding sites affected the remodeling of the chromatin.
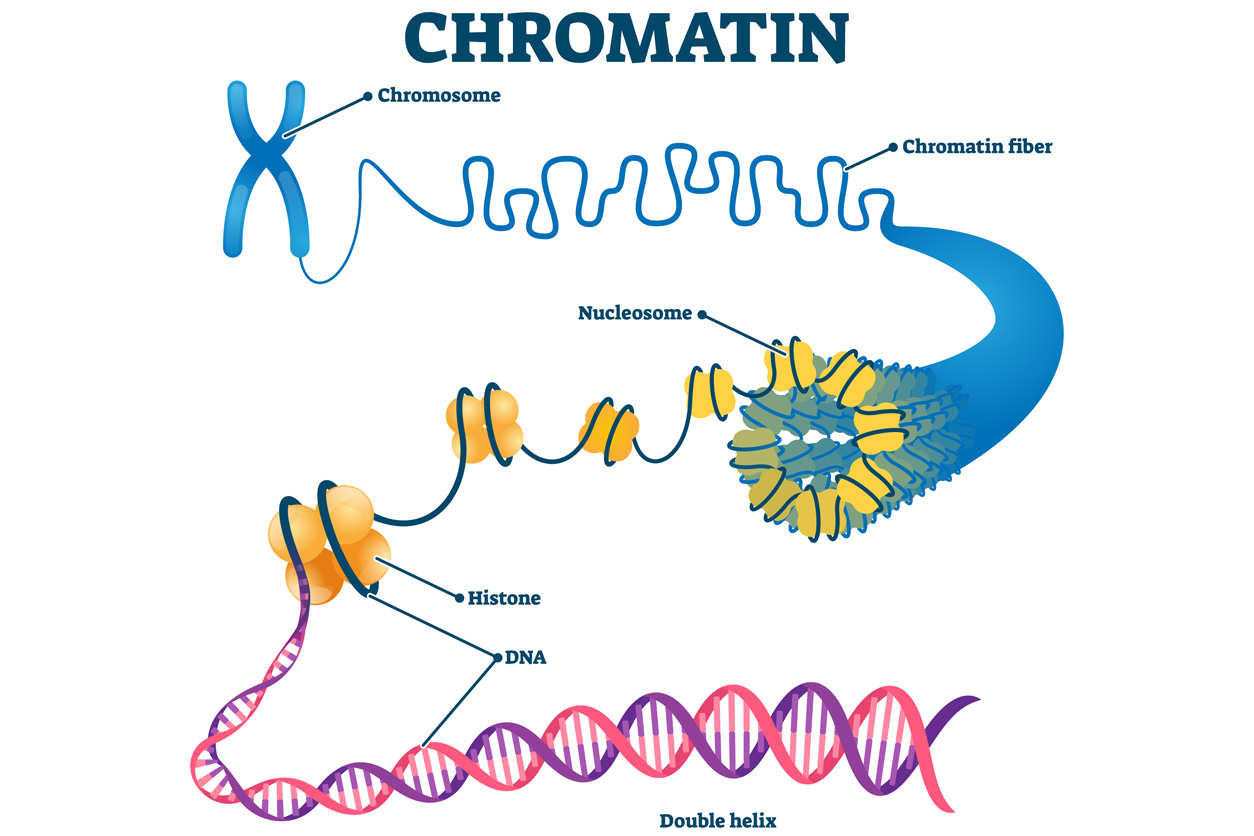 GATA3 is known as a pioneering transcription factor because it can penetrate carefully packaged chromatin fiber to access DNA and turn on genes.
GATA3 is known as a pioneering transcription factor because it can penetrate carefully packaged chromatin fiber to access DNA and turn on genes.“Frankly, I did not anticipate that the geometry of binding sites would make a difference, but it turned out to be critical for making this new light switch,” said Wade. “Our findings generate an insight into the biochemical mechanism of how this process works that is new and exciting.”
Mapping the future
Wade said he plans to use high-tech methods to map the exact geometry of the GATA3 binding sites at near-atomic resolution. Though the COVID19 pandemic cut his sabbatical short, he hopes to resume his work in Tokyo one day.
“The science will continue, but at some point we will be able to travel again and pick things up at a different spot in the story,” said Wade.
Citation: Tanaka H, Takizawa Y, Takaku M, Kato D, Kumagawa Y, Grimm SA, Wade PA, Kurumizaka H. 2020. Interaction of the pioneer transcription factor GATA3 with nucleosomes. Nat Commun 11(1):4136.
(Marla Broadfoot, Ph.D., is a contract writer for the NIEHS Office of Communications and Public Liaison.)









